fig7-new.py
About this model
| Original publication: | |
|---|---|
| Kernik et al. (2019): "A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources" J Physiol. 2019 Sep 1; 597(17): 4533-4564. | |
| DOI: | https://dx.doi.org/10.1113%2FJP277724 |
Figure 07
Slow delayed rectifier potassium current (IKs) model optimization
The voltage dependent activation gating variables were modeled here. experimental iPSC‐CM data collected from Ma et al. (2011) and two other independent datasets from Ma et al. (2015) in order to optimize data specific model. Parameters in all models were parameterized using the activation time constants from Ma et al. (2011) (Fig. 7. B).
act_inact.cellml is the main CellML file which shows the probability of transient outward potassium channel being open orclose. Its associated SED-ML file contains all the simulation settings. All the CellML files and SED-ML files need to be download in a same folder (act_inact, gating, parameter, unit) as well as python script (fig7-new.py). In the python script, required SED-ML file is loaded into the script and by running the code following figure is reproduced. fig7-new.py is used to generate the simulation and reproduces the graph shown in Figure 7 in the original study. In order to reproduce Figure 7, once all the files are downloaded to the same folder, execute the following script from the command line (command prompt):
cd [PathToThisFile]
[PathToOpenCOR]/pythonshell fig7-new.py
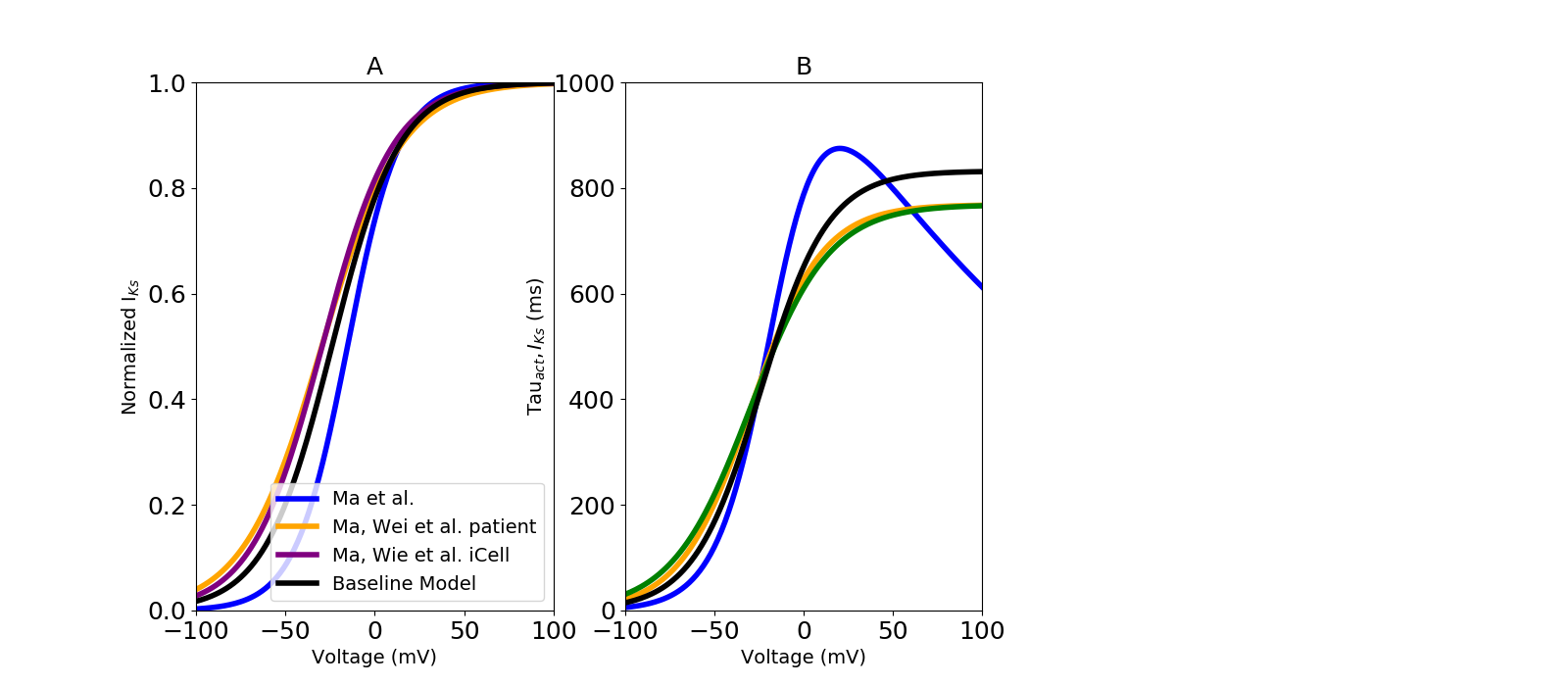
A, optimized steady-state activation with dataset-specific model fits. Different colour represent experimental iPSC-CM data from multiple laboratories. B, Time constant of IKs activation gate.
