fig3-new.py
About this model
| Original publication: | |
|---|---|
| Kernik et al. (2019): "A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources" J Physiol. 2019 Sep 1; 597(17): 4533-4564. | |
| DOI: | https://dx.doi.org/10.1113%2FJP277724 |
Figure 03
Sodium current (INa) model optimization
For sodium currents, model parameters were optimized to multiple experimental datasets, resulting in dataset-specific parameterization instances of the model. For each dataset-specific model, external ion concentrations and voltage protocols were set to reflect the corresponding experimental conditions. All of the experimental data used to optimize the models were collected in iCell iPSC-CMs. This process was used to generate dataset-specific models. Three separate experimental iPSC-CM datasets are used for sodium current: (A) Ma et al. (B) Jalife Immature, (C) Jalife Mature and (D) Baseline model
All the CellML files and SED-ML files need to be download in a same folder (act_inact, gating, parameter, unit) as well as python script (fig3-new.py). In the python script, required SED-ML file is loaded into the script and by running the code following figure is reproduced. fig3-new.py is used to generate the simulation and reproduces the graph shown in Figure 3 in the original study. In order to reproduce Figure 3, once all the files are downloaded to the same folder, execute the following script from the command line (command prompt):
cd [PathToThisFile]
[PathToOpenCOR]/pythonshell fig3-new.py
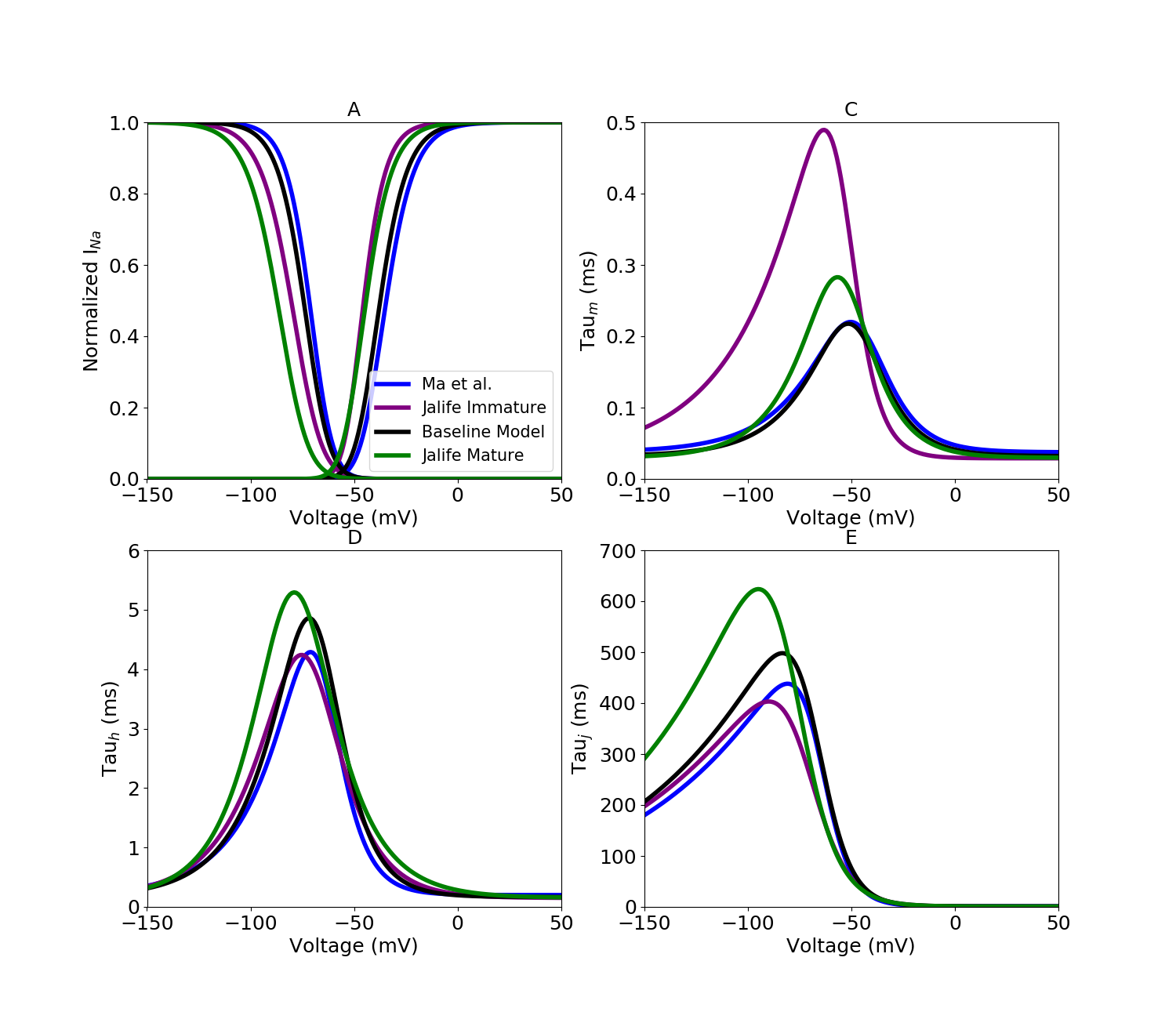
A, steady-state inactivation and activation curves. The sodium current model used in the baseline whole-cell model is shown in black. Colours represent distinct experimental iPSC-CM data from Ma et al. (2011) and from immature and mature cell preparations from the Jalife lab (Herron et al. 2016). C, INa activation (m-gate) time constants. D, INa fast-inactivation (h-gate) time constants. E, INa slow-inactivation (j-gate) time constants. j-gate time constant parameters for all INa models were optimized to experimental iPSC-CM data from the Kurokawa lab (Li et al. 2017).
