fig10-new.py
About this model
| Original publication: | |
|---|---|
| Kernik et al. (2019): "A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources" J Physiol. 2019 Sep 1; 597(17): 4533-4564. | |
| DOI: | https://dx.doi.org/10.1113%2FJP277724 |
Figure 10
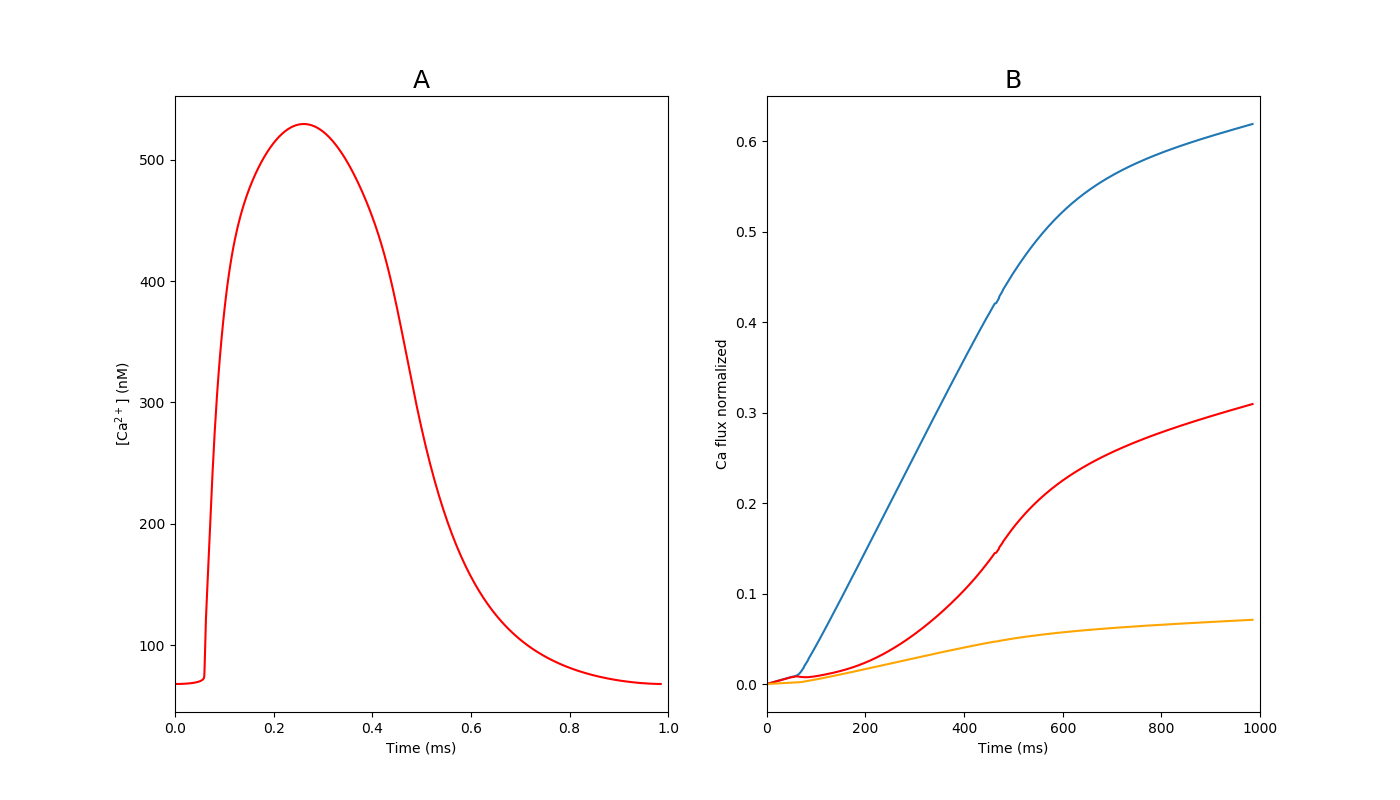
Optimization of calcium handling in the iPSC-CM baseline model
In figure 10-A baseline model of calcium transient is shown. Also the contribution of SERCA (Jup), sodium-calcium exchanger(INCX) and sarcolema pump(IPMCA) fluxes are presented in figure 10-B. comparison of the mentioned fluxes were provided by Hwang et al. (2015) from six iPSC-CM datasets within three different labs. Maximal of ISERCA, INCX and IPMCA in the baseline model first optimized to fit the relative contribution of each current during a single CaT and then normalized to total calcium contribution from all the currents for a single action potential in the baseline model after reaching steady state.
Channels.cellml is the main CellML file which shows the I-V curves for different channels fitted to different experimental data from multiple laboratories. Its associated SED-ML file contains all the simulation settings. All the CellML files and SED-ML files need to be download in a same folder as well as python scripts (fig10-new.py and ca_analysis.py) . In the fig10-new.py python script, required SED-ML file is loaded into the script and by running the code all the required currents are calculated, finally the other function in ca_analysis.py run and the following figure is reproduced. fig10-new.py and ca_analysis.py are used to generate the simulation and reproduces the graph shown in Figure 10 in the original study. In order to reproduce Figure 10, once all the files are downloaded to the same folder, execute the following script from the command line (command prompt):
cd [PathToThisFile]
[PathToOpenCOR]/pythonshell fig10-new.py