Marwan, 2003
Model Status
This is the original unchecked version of the model imported from the previous CellML model repository, 24-Jan-2006.
Model Structure
Complementation is the production of a wildtype phenotype when two recessive mutant alleles are combined in the same genotype. In fungi complementation can arise when two mutant cells form to produce a heterokaryon. In such heterokaryons two nuclear populations mix, providing the biochemical basis for the complementation effect.
In the Marwan 2003 publication described here, the author uses the photosensory control of sporulation in the protist Physarum polycephalum as an example of how complementation in a heterokaryon can create a wildtype phenotype. Marwan also investigates how such a process can depend on the time-dependent physiological state adopted after pathway activation by one of the mutant fusion partners.
The kinetic model of this process can be seen in the figure below. Two inputs, light of the far-red wavelength and glucose, control the devlopmental switch network through formation of the sporulation signal S. Photoreceptor and glucose sensor dependent pathways are integrated by a mechanism which behaves like a logic AND gate. Transcription of gene S is repressed by the product of gene V, while the transcription of gene V is reiprocally repressed by the product of gene S.
The complete original paper reference is cited below:
Theory of Time-Resolved Somatic Complementation and Its Use to Explore the Sporulation Control Network in Physarum polycephalum , Wolfgang Marwan, 2003, Genetics , 164, 105-115. (Full text (HTML) and PDF versions of the article are available on the Genetics website.) PubMed ID: 12750324
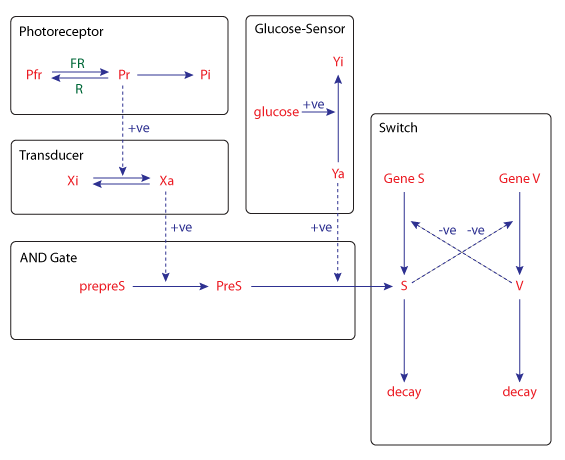 |
| Schematic diagram of the kinetic model for the sensory control of sporulation in Physarum polycephalum. |
