Model of Excitation-Contraction in a uterine SMC
About this model
| Original publication: | |
|---|---|
| Bursztyn, Limor, et al (2007): "Mathematical model of excitation-contraction in a uterine smooth muscle cell." American Journal of Physiology-Cell Physiology 292.5 (2007): C1816-C1829. | |
| DOI: | https://pubmed.ncbi.nlm.nih.gov/17267547/ |
Model status
The current CellML implementation runs in OpenCOR. The results have been validated against the data extracted from the figures in the published Bursztyn, Limor, et al (2007). We provide the settings used for the figure reproduction with the simulation results shown under Experiments. The model structure can be found in the documentation of Components. The curation process has been summarized in the Model history and Known issues.
Model overview
This workspace holds a CellML encoding of the Bursztyn, Limor, et al (2007) model. The Bursztyn, Limor, et al (2007) paper describes three Ca2 + control mechanisms: voltage-operated Ca2 + channels, Ca2 + pumps and Na + ⁄ Ca2 + exchangers, which employ the mathematical formulation proposed in Parthimos, Dimitris et al (1999). The cross-bridge model of Hai and Murphy (1988) is used to describe the processes of myosin light chain (MLC) phosphorylation and stress production, which is essentially a deterministic multi-state Markov model (MM).
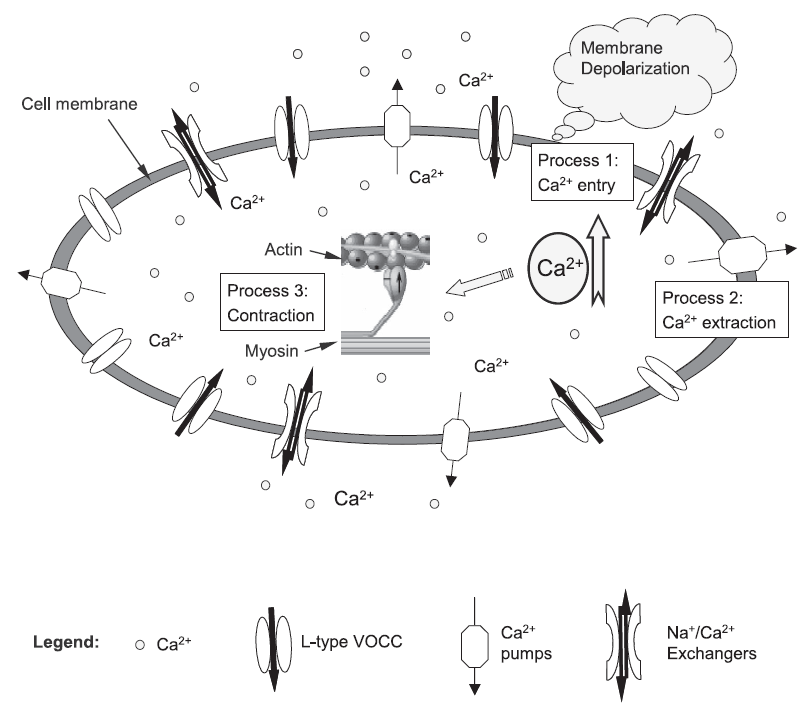
A diagrammatic representation of the Bursztyn, Limor, et al (2007) model.
Modular description
Components
CellML divides the mathematical model into distinct components, which are able to be re-used. The main CellML components are:
- Ions fluxes component, including:
- L-type voltage dependent Ca2 + channels JVOCC, and the computation of current reuses the imported ionic current components
- the efflux through Ca2 + pump JCa, pump
- the flux through the Na + ⁄ Ca2 + exchangers JNa ⁄ Ca
Excitation-contraction unit, which decouples the connection between the excitation and contraction. This is used to test individual ion fluxes and contraction development.
Each of these blocks is itself a CellML model, which enables us to reuse the various components in future studies and models.
Experiments
Following best practices, this model separates the mathematics from the parameterisation of the model. The mathematical model is imported into a specific parameterised instance in order to perform numerical simulations. The parameterisation would include defining the stimulus protocol to be applied.
This workspace has seven sets of experiments and corresponding simulation results:
Simulation settings
Simulation settings are encoded in SED-ML documents for experiment execution. It is common that we may need to vary experimental settings to obtain data under various conditions. Hence, the full experimental settings are encoded in the simulation scripts. The Python scripts to run simulation and reproduce the figures in the original paper are included under the Simulation folder. The name of the simulation and plot scripts indicates the Figure number in the primary paper. For example, Fig2_sim.py is used to generate the simulation data and Fig2_plot.py reproduces the graph shown in Figure 2 in the primary paper.
Model history
There is no publicly available code for this model.
Known issues
1. During curation process, we noticed trivial typographical errors in parameter units and references in Table 3 of Bursztyn, Limor, et al (2007). Hence, we correct these in Table 4 to remove potential confusion.
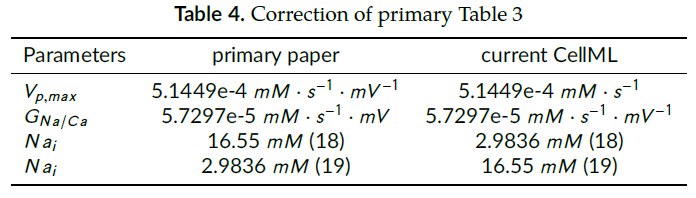
Correction of primary Table 3
- We have summarized experiment settings along with simulation results, providing more details that were not included in the primary paper.
